T clan
Description
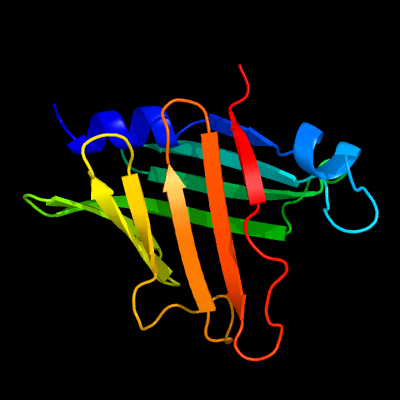
The structure of CpcT of Nostoc sp. PCC7120 and its complex with phycocyanobilin was resolved at 1.95 and 2.50 Å resolution (Protein Data Bank ID: 4O4O), respectively. CpcT forms a dimer and adopts a calyx-shaped β-barrel fold (Zhou et al., 2014). The structure of the CpeT of the cyanophage P-HM1 was resolved at 1.8 Å (Protein Data Bank ID: 5HI8; Gasper et al., 2017).
References
- Gasper et al., 2017. Distinct features of cyanophage-encoded T-type phycobiliprotein lyase Phi-CpeT: The role of auxilliary genes J Biol Chem, 292, 3089-3098.
- Zhou et al., 2014. Structure and mechanism of the phycobilin lyase CpcT J Biol Chem, 289, 26677-26689.
Showing only unsure genes
| Gene | Locus tag | Genbank | Species | Strain | PBP in rods | Pigment type | Remark | Length | Last change |
|---|---|---|---|---|---|---|---|---|---|
| CpcT family | GTHECHR1104 | XP_001713602.1 | Guillardia theta | CCMP2712 | Cr-PE 545 | n.a. | Low protoscan score but best BLASTP match to the CpcT family. This cpcT-like gene was characterized by complementation of Synechocystis sp. PCC6803 cpcT (Bolte et al. 2008). However, since G. theta possesses PE (Cr-PE 545) not PC, the function of this gene (despite its closest similarity to cpcT than cpeT families) must likely be to attach a PEB at Cys-159 of beta-PE. | 222 | 09-28-2012 |
| CpcT family | CMK263C | Cyanidioschyzon merolae | 10D | PC | 1 | found by tblastn | 263 | 02-21-2012 | |
| CpcT family | HAN_1g134 | XP_001712298.1 | Hemiselmis andersenii | CCMP644 | Cr-PE 555 | n.a. | 221 | 01-24-2012 | |
| CpcT family | Pse7367_2699 | YP_007103382.1 | Pseudanabaena sp. | PCC7367 | PC+PEI | Low protomatch score | 202 | 06-20-2013 | |
| CpeT family | CwatDRAFT_5720 | ZP_00514725.1 | Crocosphaera watsonii | WH8501 | PC+PEI | Similar to CpeT, but with a 51 aa deletion in the middle. N- and C-term closely related to WH0003 cpeT sequence. | 149 | 09-26-2012 | |
| T homologs family | Syn6312_3060 | YP_007062655.1 | Synechococcus sp. | PCC6312 | PC | 192 | 06-12-2013 |
![]() Model strains, Marine strains
Model strains, Marine strains
Last update: 17 Jan 2018 18:31:49